-
October 2020: Team "MSFragger Swagger" wins the #ALSMinePTM Challenge!
Kevin (top center) led the team to victory in identifying protein post-translational modifications (PTMs) related to ALS. Team MSFragger Swagger: Fengchao, Danny, Hui-Yin, Dan, and Sarah. Read more at News in Proteomics Research.

-
June 2020: Check out our presentations at the ASMS Reboot
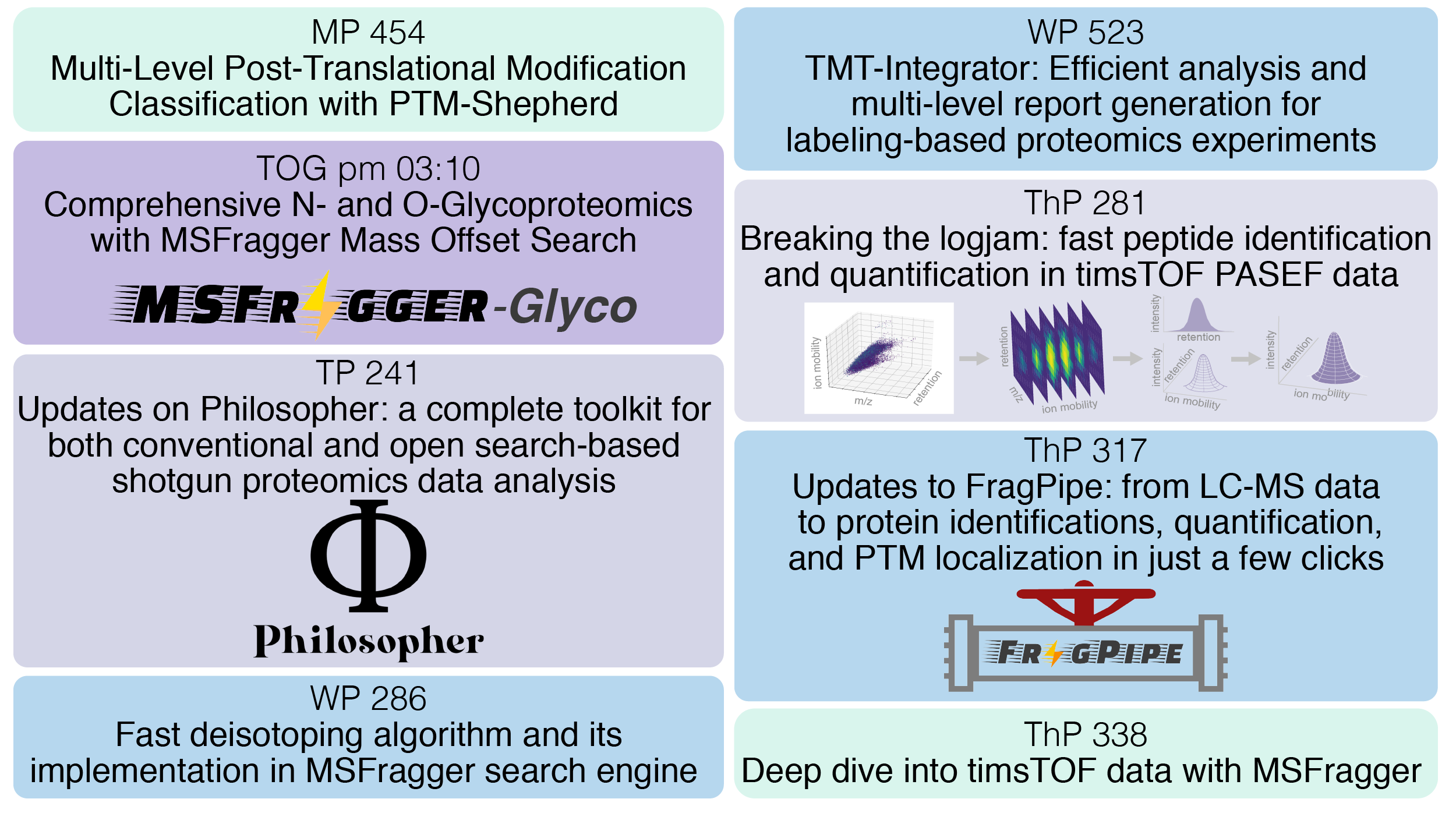
-
May 2020
Welcome Kevin!
Welcome to our new Ph.D. student Kevin! We are excited Kevin Yang is joining us for his doctoral studies.
-
November 12, 2019: Snow day in November! (missing Dmitry)

-
July 01, 2019
Welcome Dan!
Welcome to new post-doctoral fellow Dan A. Polasky. Dan joins us from the University of Michigan Chemistry Department. Dan received his Ph.D. in Chemistry.
-
June 4, 2019
Lab dinner at ASMS in Atlanta!

-
May 31, 2019
Check our posters at ASMS
If you are going to attend the annual meeting of the American Society for Mass Spectrometry (ASMS) in Atlanta, make sure to stop by at our posters:
- MP 402 Comparison of Open-Search Tools.
- MP 405 Shifted Ions Searching and Other Improvements in the MSFragger Database Search Engine.
- MP 416 FragPipe: A Fast Proteomics Pipeline with MSFragger Search Engine at Heart.
- MP 437 The implementation of MSFragger and Philosopher (PeptideProphet) nodes in Proteome Discoverer.
- WP 396 Updates on Philosopher: a complete toolkit for both conventional and open search-based shotgun proteomics data analysis.
- ThP 693 Exploring the Open Proteome: Proteomics Open Search Analysis with PTM-Shepherd.
-
May 01, 2019
Welcome Sarah!
Welcome to new post-doctoral fellow Sarah E. Haynes. Sarah joins us from the University of Michigan Chemistry Department. Sarah received her Ph.D. in Chemistry.
-
January 01, 2019
Welcome Fengchao!
Welcome to new post-doctoral fellow Fengchao Yu. Fengchao joins us from the Hong Kong University of Science and Technology, Division of Biomedical Engineering. Fengchao received his Ph.D. in Bioengineering.